Everyone Agrees: Let's Put Genetics to Work
It's time already.
When two scientific papers disagree, controversy invariably ensues — unless papers disagree in the same direction.
That’s what happened with Nelson et al. (2015) and King, Davis & Degner (2019).
Nelson et al. found that when drugs had genetic support — for example, because genes coded the drug target — they were more likely to achieve approval. With musculoskeletal drugs, metabolic drugs, neurological and behavioral drugs, cancer drugs, etc., genetically-supported ones were more successful. And the effect was large! Drugs with genetic support were apparently twice as likely to receive approval.
King, Davis & Degner updated Nelson et al.’s dataset and produced revised estimates of the effect of genetic evidence. Now, instead of a doubling of the approval chance, genetic support is sometimes consistent with up to an octupling of the approval rate.
This all makes sense for a lot of reasons. We know that phenotypes associated with drug targets predict side effects in clinical trials, and we know that GWAS hits have modest effect sizes, but the minuteness of their effects obviously can’t preclude a far larger therapeutic impact via their targets. A commonly cited example is variants in the HMG-CoA Reductase (HMGCR) gene. The HMGCR gene codes an enzyme that statins aim to reduce to treat dyslipidemia1. The variants in the HMGCR gene have individually meager effects, but if we took all the risk alleles, it would theoretically be possible to calculate the consequences for the dyslipidemia phenotype if we reduce or increase the activity of the enzyme that gene produces. This effectively means that therapeutic calibration is on the table once you have a sufficient, and indeed, achievable, level of genetic knowledge.2
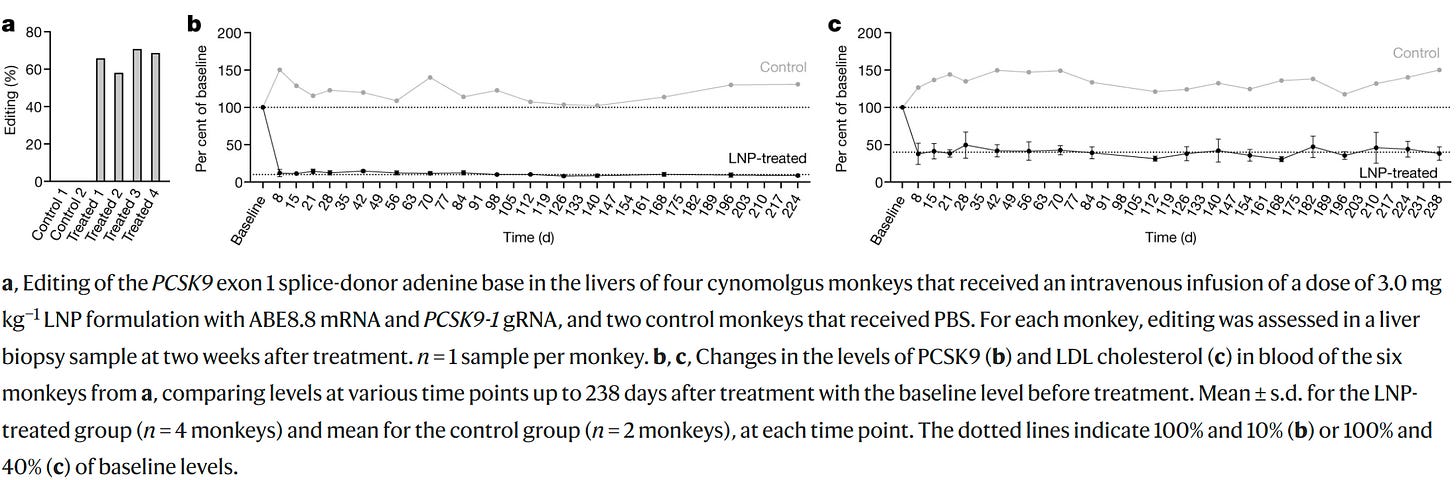
But even if we don’t do this, just knowing the target makes for highly effective drugs, even though the variants in the GWAS don’t do much. The difference in dose between the genetic effect and the therapeutic one can be incredible, like the difference between being shot with a BB and being shot with .50 BMG. And in some cases, knocking out a gene like PCSK9 can have a much larger effect on a person or animal than you might expect given small variant or genic effect sizes. This example is real, just take a look at what happens when macaques are edited to cause almost complete knockdown of the gene:
Genes being used to find drugs and yield therapies for various ailments is one of the more anodyne applications for the results of the GWAS revolution. What if we go up an offensiveness level? Vassy et al. (2023) have some new data that let’s us do just that.
Vassy et al. conducted a national survey of physicians to figure out what they thought about using polygenic risk scores (PRS)3 with their patients. Doctors tended to think pretty highly of using PRS. They considered them useful for identifying patients who need preventative medication, for boosting the efficacy of screening procedures, and for recommending lifestyle modifications.
And when it came to patient and provider decision-making as well as the idea that PRS could improve patient health outcomes, they were also in agreement: PRS look promising.
And there’s only really modest or misguided concern about barriers to use:
The notion that some of these are misguided isn’t meant to be insulting, it’s just realistic: PRS can be explained, research in non-White people can increase and, indeed, is increasing, PCPs can figure out how new technology that can help their patients works, PRS should reduce the number of unnecessary tests and treatments, patients are anxious about many medical things when they’re new and old but that can decline with time and shouldn’t get in the way of operating, details can be hidden from insurers by legislative fiat, clinical guidelines can be developed, and out of pocket costs are actually low and will continue to fall.
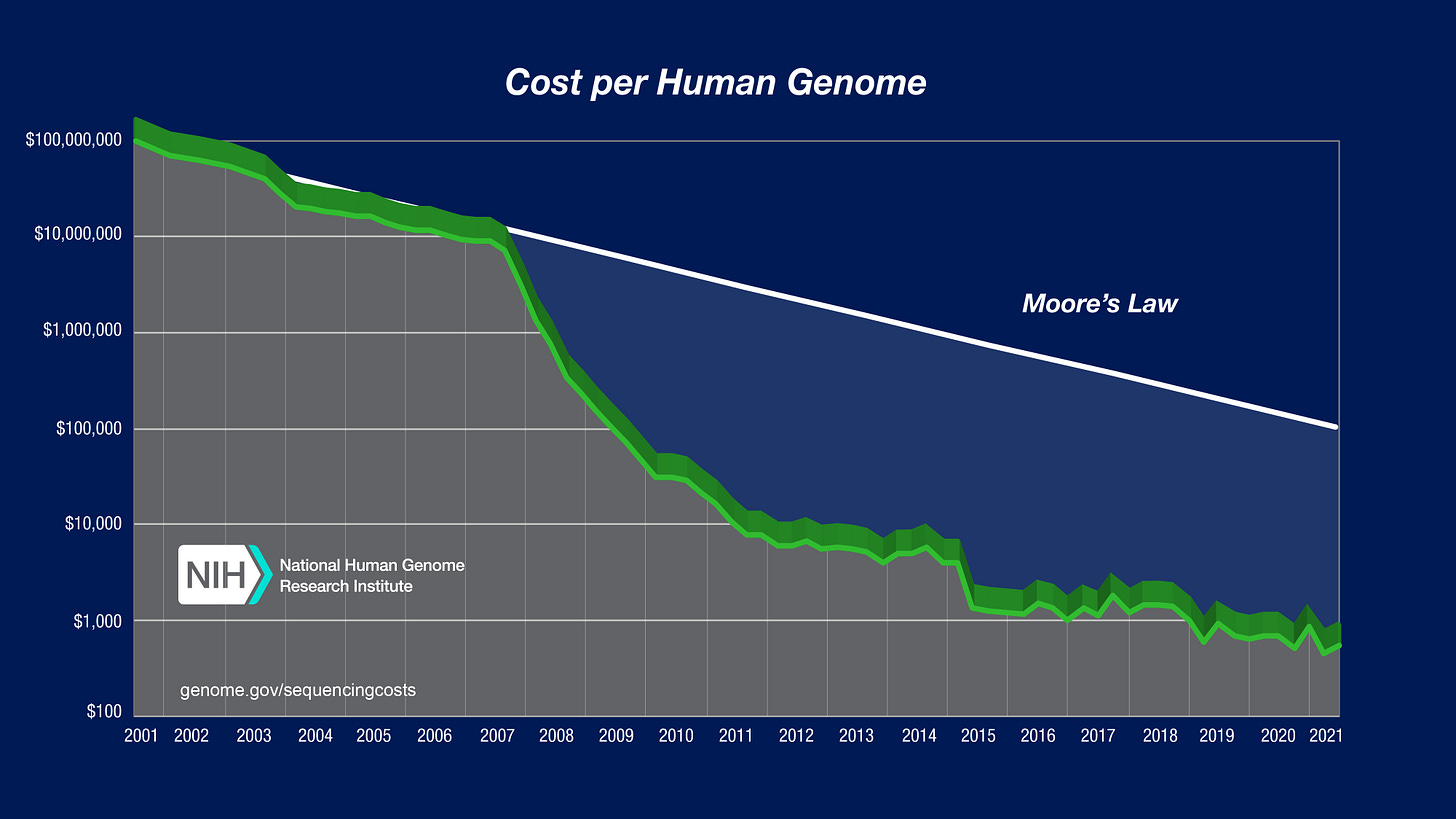
The last part is one I want to focus on because, now that it costs $250 to get a whole genome sequence with 30x coverage from a company like Nebula Genomics, there’s really no excuse for not getting every patient sequenced. That cost of sequencing is way down from even the NIH’s Moore’s Law-beating 2021 numbers:
Physicians who are aware of this area at all may, rightly, think it’s a lot more expensive to start using than it really is. They might have seen products like Myriad’s MyRisk that are much more expensive than and inferior to Nebula’s whole genome sequencing4 and gotten the wrong impression of genetic testing thanks to these outdated and overpriced products. They may be concerned that, even if they get someone's whole genome sequence data, they won't have the tools to generate the PRS they need. It really only takes a few seconds to generate a person's various PRS, so this concern could be met by some enterprising young entrepreneur with relative ease.
Back to utility, doctors do have the right idea. PRS work in clinical settings.
While monogenics dominate our knowledge of conditions like breast cancer, polygenic screening is well on its way, and could immediately afford utility for predicting disease subtypes like luminal A-like and triple-negative disease.
Fracture risk assessment tools are widely used in the management of osteoporosis. Unfortunately, despite being quick and easy to use, they don’t offer doctors much more information than age, sex, and BMI. Fortunately, more complex assessments have heritable results, and so PRS for the results of those assessments can be computed and used to predict other patient’s fracture risk from their genetic data.
Some conditions like gout and prostate cancer are caught early by testing condition-relevant biomarkers like uric acid and prostate-specific antigen levels. For a subset of the population, biomarker readings don’t seem to work for identifying the presence of these conditions. Genetic predictors on the other hand will much more reliably indicate risk. Take prostate cancer as an operative example. A recent paper by Ma et al. (2023) provided details of a potentially lifesaving use of PRS to identify men at risk of developing prostate cancer despite low prostate-specific antigen levels. By using PRS, they may have created an opportunity to find potential prostate cancer sufferers long before cancer cropped up, giving them ample headway to alleviate people’s suffering and save their lives.
Upper gastrointestinal cancer has numerous risk factors, but most important are diet and genes. By understanding individual’s genetic risk, doctors can more forcefully recommend dietary changes that might help to mitigate the impacts of genetic background and prevent upper gastrointestinal cancer from ever appearing among those most at risk.
In areas where monogenic screening is already heavily used like with atrial fibrillation and risk of breast cancer, PRS are catching up or are already ahead, so in as much as monogenic screening is in common use, polygenic screening should be too. Consider the case of venous thromboembolism: people with a high value for the PRS are as at risk as people with known, powerful monogenic causal variants.
Lu et al.’s (2022) work also bolsters the cost-effectiveness angle for polygenic screening:
Monogenic causes of complex diseases were more prevalent among individuals with a parental disease history than in the rest of the population. Polygenic risk scores showed moderate discriminative power to identify familial monogenic causes. For instance, we showed that prescreening the patients with a polygenic risk score for type 2 diabetes can prioritize individuals to undergo diagnostic sequencing for monogenic diabetes variants and reduce needs for such sequencing by up to 37%. Among individuals with a family history of complex diseases, those with a low polygenic risk score are more likely to have monogenic causes of the disease and could be prioritized to undergo genetic testing.
And lest we forget, the more PRS there are and the better they are, the more drug targets the data will indicate, the more treatments will become available via finding new drugs and indicating opportunities for novel gene therapies.
If we really want to get serious about health and doctors can manage to cooperate, they can bolster efforts to find new drugs and treatments by allowing patients to opt in to letting researchers use their data for the greater good. They have the possibility to figure out genetic interactions with drugs that produce certain side effects and to learn whether certain genotypes dispose people to greater likelihoods of success with certain drugs compared to others. If doctors pursue this wholeheartedly, they can help to reduce the number of drugs and dosages psychiatric patients have to cycle through before finding what works for them and they can help to prevent potentially life endangering side effects from ever rearing their heads.
What if we up climb one more rung up the ladder of controversy? Meyer et al. (2023) have provided some data to allow us to do just that.
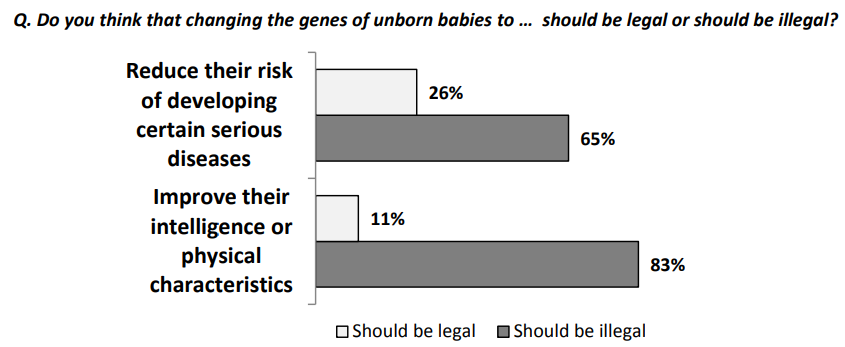
This research group bravely asked: what does the public think about the polygenic screening of embryos? This technology is eugenic, plain and simple. It lets parents choose which of a set of embryos they want to implant for whatever reasons. They could want the smartest child of a batch, the tallest child their seed can produce, or they might just want what most parents want, which is the healthiest possible kiddo.
The public is much warmer than one might naïvely expect when it comes to eugenic technology. Let’s first look at how the public views the morality of embryo selection.
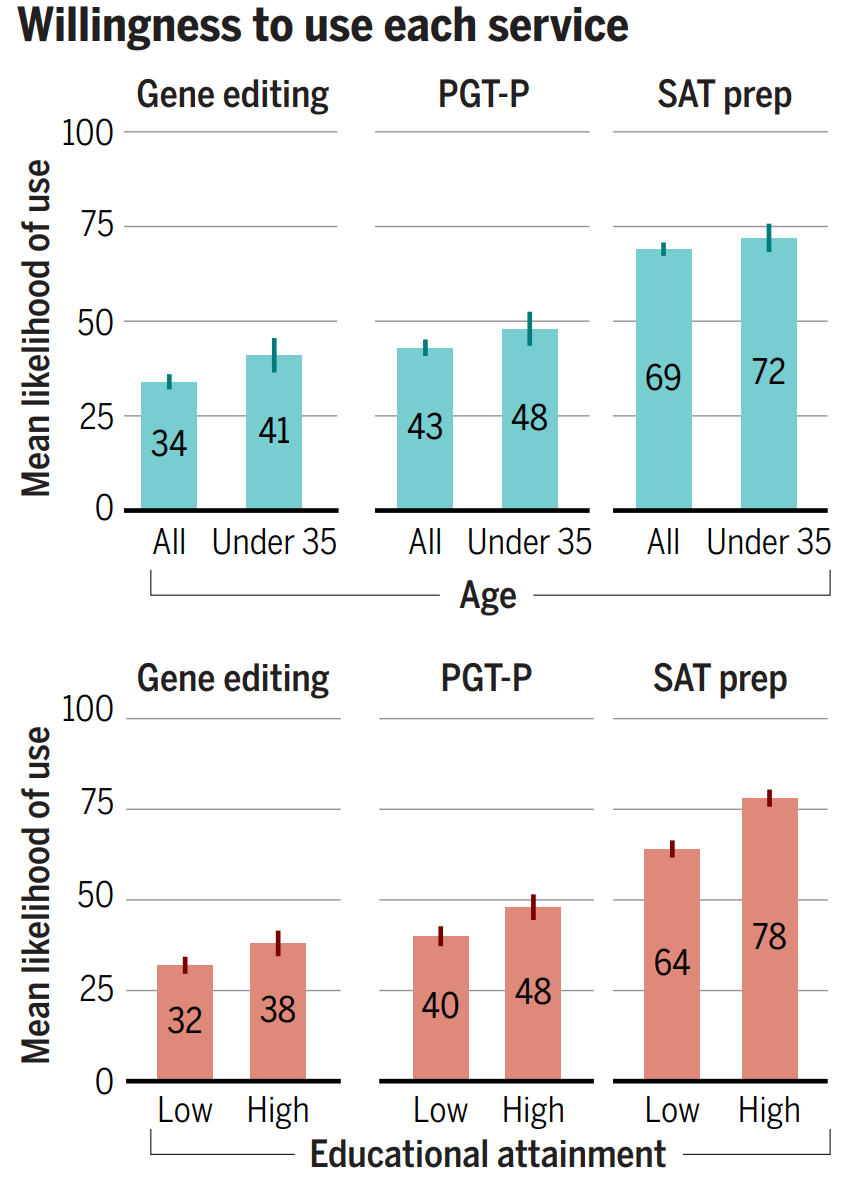
17% of the poorly-educated and 17% of highly-educated think polygenic embryo screening (PGT-P) is immoral, but this compares favorably, given that 54% of the poorly-educated and 65% of the highly-educated consider it to either not be a moral issue or to be morally permissible. The remainder may not even disapprove, since they’re people who really just aren’t sure. This isn’t much worse than the public perception of something so benign as SAT prep, and with time and people born with the method, perceptions will probably move in the same direction like they did with IVF in general so long ago.
Compare embryo screening with gene editing, and the story still looks good. Only 31% of the highly-educated and 29% of the poorly-educated think gene editing is outright wrong. If they’re confronted with a child who has Huntington’s chorea or thalassemia, presumably they’ll end up like the Arnold clone in The 6th Day and they’ll come to accept it. If the same happens to their friends, perceptions will gain the opportunity to change in a contagious way. It’s really just a matter of time, and it’s lucky that so many are either ambivalent or accepting of the technologies as they are.
What about individual’s likelihoods of actually using these services for themselves or their families?
Nearly half of highly-educated people are already willing to use PGT-P, and only about 10% fewer are willing to use gene editing outright. I highly recommend perusing the paper’s supplementary materials if you want to learn more.
The situation is exceptionally bright in this field of research, and clearly genetic prediction and sequencing are both ready to be put to use. Perhaps even more importantly though, the public has shown that it’s ready for them.
The opportunities available to us in this day and age are beyond incredible, and with the development of new technologies like in-vitro gametogenesis and iterated embryo selection or improved gene therapies and new drugs or novel methods of gene and drug discovery to supplement them all, we’re on the cusp of a golden age.
Updates
April 17: The Global Times reported on a 2018 survey out of Guangdong, Hong Kong, and Macao. The survey showed that the majority of those place’s general public and people living with HIV/AIDS supported some amount of gene editing, although support varied considerably by the targeted trait. For example, less than 15% of either category supported editing skin color, but more than 70% of the general public and almost all of those living with HIV/AIDS welcomed gene therapy for HIV prevention.
And as regards clinical use, here’s what the sample thought:
In January 2016, a Harvard poll about gene editing came out, and the public looked less supportive back then. Here are some of their choice responses:
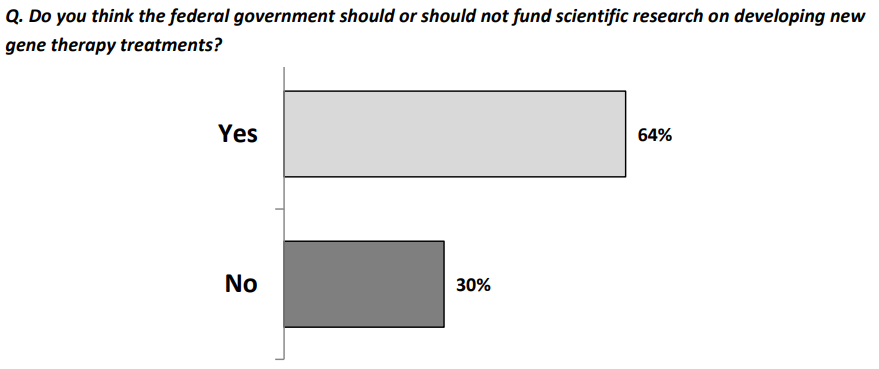
Despite these seemingly dismal views, they wanted gene therapies approved:
and they wanted research funded:
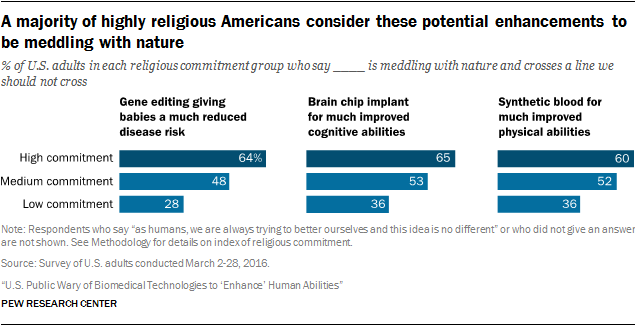
Pew conducted some polls and wrote two articles on the topic in 2016 and the results were fairly typical. What’s worth noting is the moderation of acceptance by religiosity, where more religious Americans were less likely to accept gene editing.
In one of the posts, Pew showed that people disliked gene therapy and other technologies more the better they were for the people who benefitted from them:
In 2018, Pew ran another poll on gene therapy, and views by religious status seemed to polarize more.
This post will be updated as I become aware of additional polls.
High fat content in the blood in the form of triglycerides, cholesterol, or other types of fat.
Assuming equivalence between genetic and therapeutic effects, which cannot always be safely assumed because drug targets can do multiple things and developmental effects may cause gene and therapeutic effects to deviate from one another. Figuring out if this is the case is an empirical question.
Genetic predictors of various conditions and traits. They are also known as genome-wide polygenic scores (GPS) and polygenic scores (PGS).
In the case of Myriad, only 48 genes are covered, and most of them to lower coverage than what Nebula offers for the whole genome.